

CompASS is a tool that matches the secondary structure elements of a protein with the secondary structure elements
of the same protein in a different state given their atomic coordinates. Users can submit the coordinate files (pdb files) of the protein along
with a name to make their identification easier in results.
The assignment of secondary structure is based on STRIDE (Structural identification), an algorithm for the assignment of protein secondary structure elements given the atomic coordinates of the protein. The assigned conformations are : Alpha helix, 3-10 helix, PI-helix, Extended conformation, Isolated bridge, Turn and Coil.
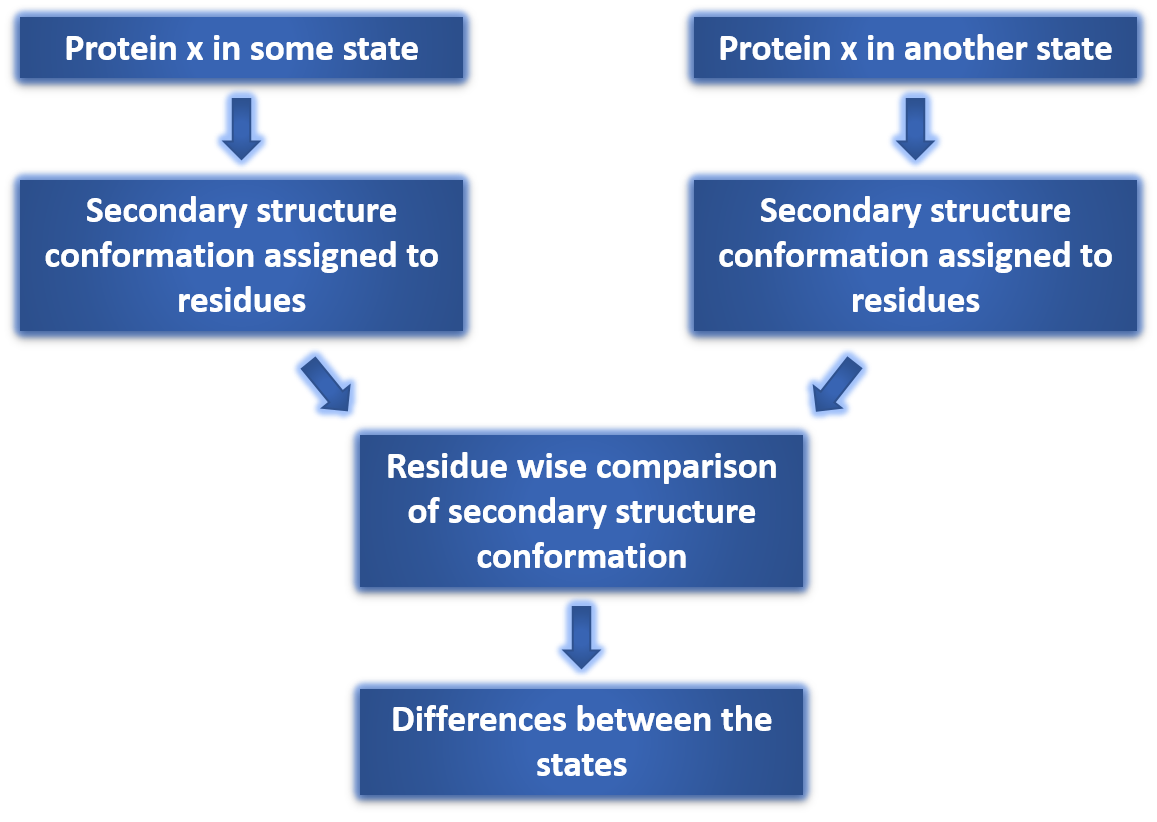

Differing Score is the percentage of residues that have different secondary structure conformation in the given two states of protein. Residues that differ in conformation are represented by " * " in alignment.
The content percentage of individual secondary structure conformation (Alpha helix, 3-10 helix, PI-helix, Extended conformation, Isolated bridge, Turn and Coil) in both the states and the difference between them is also given.
| Copyright Ⓒ 2019 Ayaluru Murali | Ankur Chaurasia | All Rights Reserved | Department of Bioinformatics | Pondicherry University |
| Header background image by David Zydd, Webpage designing learned from w3schools. |