


PreSSM is the web interface of a tool that matches the predicted secondary structure in a protein given it's sequence with the
secondary structure in the protein's 3D model given it's atomic coordinates. Users can get the prediction of secondary structure
from either of PSIPRED, CFSSP and NSPA servers. The assignment of secondary structure is based on STRIDE (Structural identification),
an algorithm for the assignment of protein secondary structure elements given the atomic coordinates of the protein.
PSI-blast based secondary structure PREDiction (PSIPRED) is a method used to predict protein secondary structure. It uses artificial neural network machine learning methods in its algorithm. CFSSP server predicts secondary structure of protein from the amino acid sequence by implementing Chou & Fasman algorithm.
At NSPA server, methods using different approaches are combined to predict secondary structure. The first step there is to select the secondary structure prediction methods (given below) you want to use. Then, the secondary structure consensus prediction program generates a secondary consensus. In the consensus the most present predicted conformational state is reported for each residue.
Secondary structure prediction methods available at NSPA server are :
DPM, DSC, GORI, GORIII, GORIV, HNN, MLRC, PHD, PREDATOR, SIMPA96, SOPM, SOPMA.
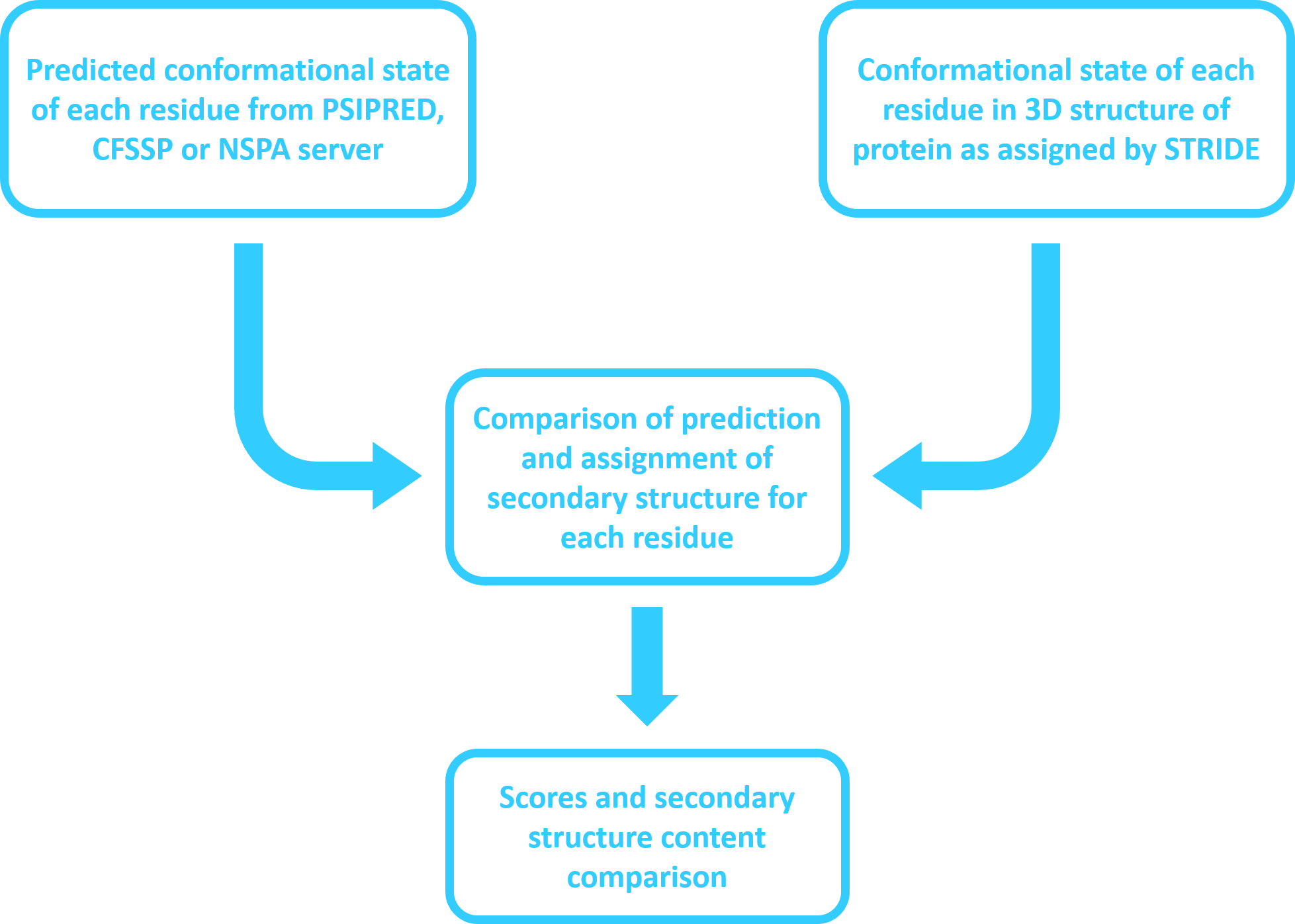

Match score is the percentage of residues that have the same conformational state given in prediction as well as in 3D model secondary structure assignment. A match is represented by " * " in alignment. For a good agreement between prediction and assignment, match score should be high
Contradiction score is the percentage of residues that have helix conformational state in prediction while sheet conformational state in 3D model secondary structure assignment or or vice versa. A contradiction is represented by " | " in alignment. For a good agreement between prediction and assignment, contradiction score should be low.
Helix, Sheet and Coil content is the percentage of residues in respective conformational states in prediction as well as in 3D model secondary structure assignment. A difference between these percentage is also given to observe deviation between prediction and assignment.
| Copyright Ⓒ 2019 Ayaluru Murali | Ankur Chaurasia | All Rights Reserved | Department of Bioinformatics | Pondicherry University |
| Header background image by David Zydd, Webpage designing learned from w3schools. |